Connection Parameters
These parameters are usually imported from "_geom_bond_xxx", "_geom_contact_xxx", and "_geom_hbond_xxx" loops in CIF
Connection parameters in brief:
- Parameters are editable in the dialog "Structure/Connection Parameters", which
has three tabs (pages).
- Tables of bond, H-bond, and contact parameters are available from the "Tables"
sub-menu of the "View" menu.
Previous article: Atomic parameters
Next article: Automatic vs. manual picture creation
We use the compound COD:1501635 for demonstation in this article. Opening the sample
file COD-1501635.diamdoc starts with a blank picture.
Bond, H-bond and contact parameters
You have been knowing the table of atomic parameters in the data pane right beneath
the structure picture view since Diamond version 2. Since version 4 this table is supplemented
by three more tables:
a) The table of bond parameters. These parameters are usually derived from CIF "_geom_bond_xxx"
loops and define selected bonds (that may be important in a publication for instance)
or even a whole (ensemble of) molecule(s).
b) The table of contact parameters. Like the bond parameters, these usually come
from "_geom_contact_xxx" loops.
c) The table of H-bond parameters. These describe relations between an H-atom, a
donor atom and an acceptor atom each with bond lengths/atom distances and an angle
each (usually from "_geom_hbond_xxx").
Like the atomic parameters, these connection parameters can be edited using the
command "Connection Parameters..." from the "Structure" menu. The dialog has three
tabs (pages) for the bonding, contacting and H-bond parameters, rsp.
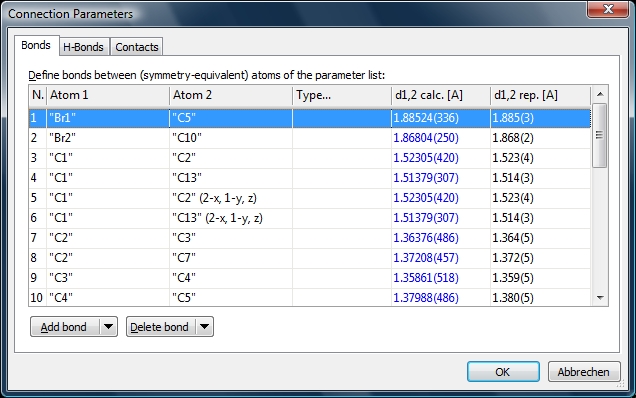
Screenshot of "Bonds" page of "Connection Parameters" dialog listing bond parameters
defined in COD-1501635.diamdoc. The rightmost column shows the reported distance
(usually from _geom_bond_distance in CIF loop), "d1,2 calc." the value calculated
by Diamond.
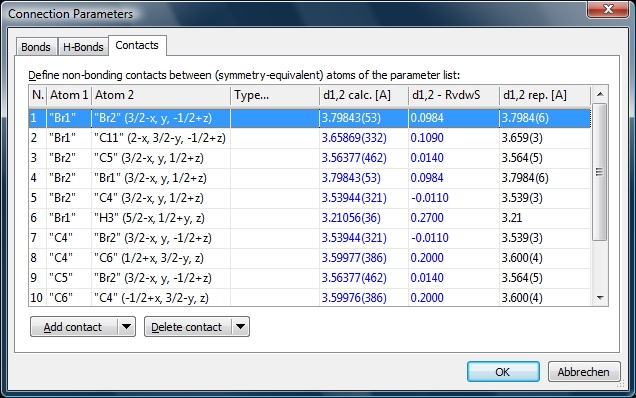
Screenshot of "Contacts" page of "Connection Parameters" dialog listing parameters
for non-bonding contacts, defined in COD-1501635.diamdoc. The rightmost column shows
the reported distance (usually from _geom_bond_distance in CIF loop). "d1,2 calc."
is calculated by Diamond. "d1,2 - RvdwS" is the distance relative to the sum of
the van der Waals radii.
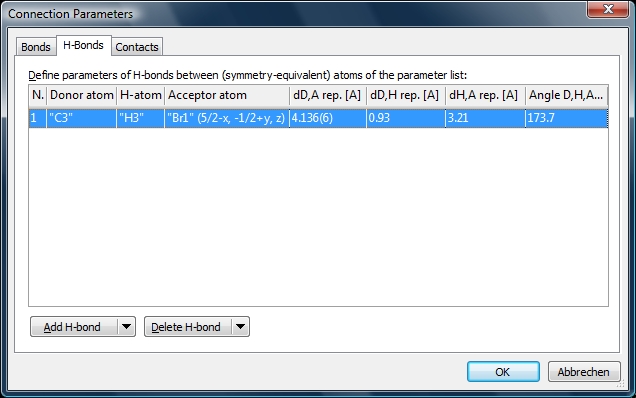
Screenshot of "H-Bonds" page of "Connection Parameters" dialog listing H-bond parameters
defined in COD-1501635.diamdoc. The four numeric values shown are the distance between
donor and acceptor, between donor and H, between H and acceptor, as well as the
angle D,H,A, reported usually from _geom_hbond_xxx CIF items.
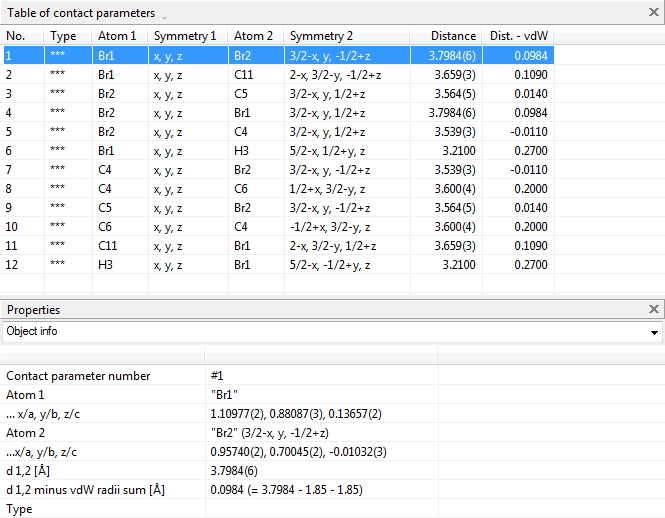
Tables of bond, contact, and H-bond parameters
In addition to the table of atomic parameters, the tables for bond, H-bond, and
contact parameters are available through the "View"/"Table" sub-menu or from the corresponding
dropdown menu in the right part of the main toolbar. The screenshot shows COD:1501635
with table of contact parameters and properties of the highlighted
contact parameter:
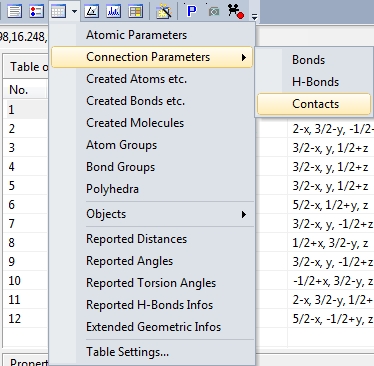
Screenshot of "Table" dropdown menu from the main toolbar
Adding all atoms (and connections) in a structure picture
In some cases, the bond parameters are used to strike out several bonding spheres,
spheres that may differ from those generated by effective radii, for instance. But
in many other cases, the bond parameters, together with the atomic parameters, can
serve as a simple starting point to create molecule(s) directly from the atomic
parameter list. This is done in Diamond with the command "Add All Atoms and Connections..."
from the "Build" menu. This also considers non-bonding contacts and H-bonds when mentioned
in the structure input parameters. Read the article "Adding All Atoms (and Bonds) of the Parameter List(s)" for details.
Previous article: Atomic parameters
Next article: Automatic vs. manual picture creation
References:
COD:1501635: Ulrich Darbost, Janie Cabana, Eric Demers, Thierry
Maris, James D. Wuest; Molecular Tectonics. "Use of Br...aryl Supramolecular Interactions
for the construction of Organized Networks from 9,9'-spirobifluorene in the Crystalline
State"; CheM, 1 (2011), 52-12369.
|

